Multi-Organ Segmentation
Multi-Organ Segmentation using Deep Neural Networks
IntroductionIn recent times, Deep Leaning (DL) has gained more attention in different domains, which includes, Computer Vision tasks, Image Processing, Autonomous Driving, and in Machine Vision applications. Complex Imaging tasks have been addressed with the help of Deep Convolutional Neural Networks (CNNs) by training on large datasets to predict the class, segment the region of interest, and to detect the changes over the time efficiently. The CNNs learn key features from the training data over the time. As a result, researchers have explored the use of CNN based methods on other imaging domains such as Medical Image Analysis and Segmentation, where better feature representation and precision are the most important factors in image analysis. However, more attention is needed towards Medical Imaging domain as the manual segmentation of a medical image is time consuming, which results in high cost, more delay in diagnosis, and misdiagnosis. In this research, we focus on automatic segmentation of the pancreas in computed tomography (CT) images, that helps the radiologists in assisting to identify the pancreatic tissue easily and diagnosis more patients in short amount of time. Automatic segmentation of Pancreas is a hard problem because of its high variability in shape, size and location [1]. Most of the time, the target occupies a small region (e.g., <0.5%) of the entire CT scan and less contrast variability between the region of interest and the background. As a result, the deep CNNs, such as FCN and other networks, find it difficult to segment the region of interest accurately. Therefore, this work is mostly concentrated on improving the automatic pancreas segmentation accuracy by using the Recurrent Residual U Net (R2U-Net) model [2]. Figure 1 shows a typical example from the National Institute of Health (NIH) dataset in three different views from left to right (x, y and z -axis).
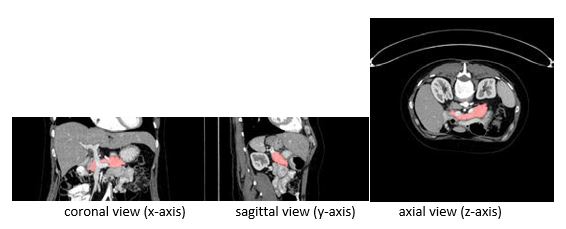 Figure 1. An example from the NIH pancreas segmentation dataset visualized in three views.
Figure 1. An example from the NIH pancreas segmentation dataset visualized in three views.
Goals/Objectives
- Accurate segmentation of the pancreas region in complex cases.
- Design an end-to-end coarse-to-fine multi-organ segmentation model.
- Build the model in generalized way so that it can adapt to segment multiple organs.
Methodology
To segment the pancreas region with high accuracy, the architecture deals with multiple stages of training, called as coarse-to-fine [3] approach. The coarse and fine stages are trained with same R2U-Net model. The first stage (coarse-scaled model) takes the entire slice as an input and uses it to predict the initial region of the pancreas. The second stage (fine-scaled model) is only trained on the exact pancreas region by eliminating the background noise for accurate segmentation around the pancreas region. Figure 2 shows the flow chart of the entire architecture.
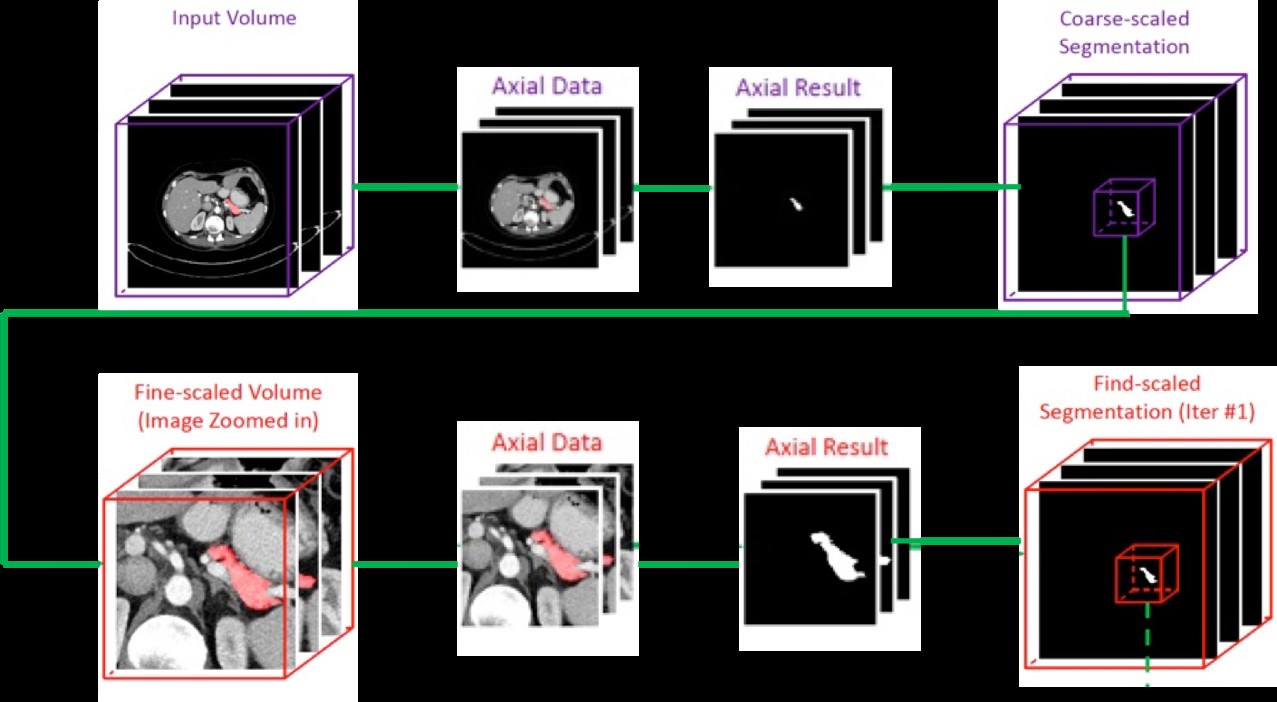 Figure 2. Architecture of the coarse-to-fine models. The top row represents the coarse-scaled model and bottom row represents the fine-scaled model setup.
Figure 2. Architecture of the coarse-to-fine models. The top row represents the coarse-scaled model and bottom row represents the fine-scaled model setup.
R2U-Net Architecture
To train the coarse and fine models, we used the R2U-Net architecture instead of the regular Convolutional Neural Networks (CNNs). The major advantages in using the R2U-Net is that the residual units are added with the forward model in the encoding and decoding units, which helps in training deep networks. Recurrent residual convolutions with multiple time steps are used for better feature representation for segmentation tasks. Figure 3 shows the architecture of the R2U-Net model and we refer to the original paper [2] to understand more about the model.
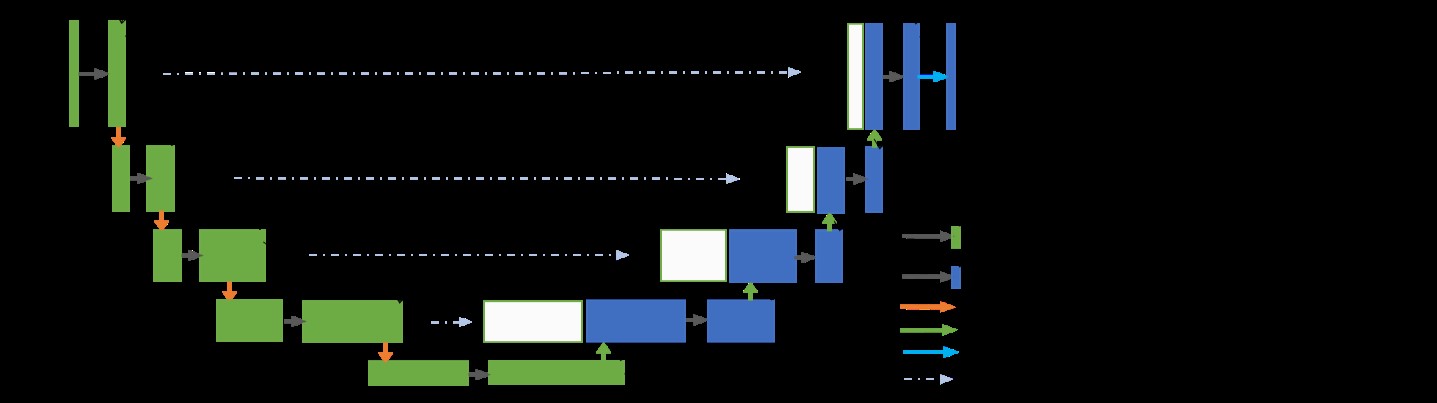 Figure 3. Architecture of the Recurrent Residual U-Net (R2U-Net) model.
Figure 3. Architecture of the Recurrent Residual U-Net (R2U-Net) model.
Results/Evaluations
The NIH pancreas segmentation dataset [1] is used to evaluate the model, which contains 82 contrast-enhanced abdominal CT scans. The database subjects were normal and have neither major abdominal pathologies nor pancreatic cancer lesions. Their ages range from 18 to 76 years with a mean age range from 46.8 to 52.4. The resolution of each CT scan is 512x512xL, where L [181, 466] is the number of sampling slices along the z-axis. The slice thickness varies from 0.5mm to 1.0mm. We followed the standard cross-validation strategy, split the dataset into 4 fixed folds, each contains 20 cases. We trained the model on 3 out of 4 folds and tested on the remaining fold by measuring the Dice-Sorensen Coefficient (DSC) for each case. 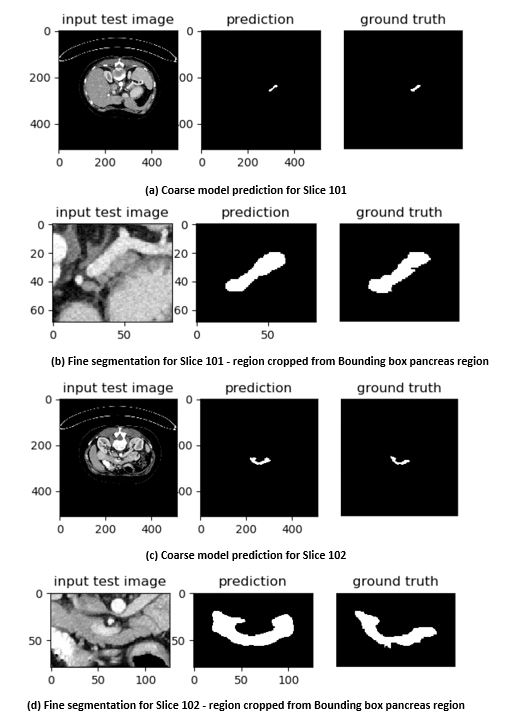 Figure 4. Case #001 (Slices 101 and 102) showing inter slice variations in a single case.
Figure 4. Case #001 (Slices 101 and 102) showing inter slice variations in a single case.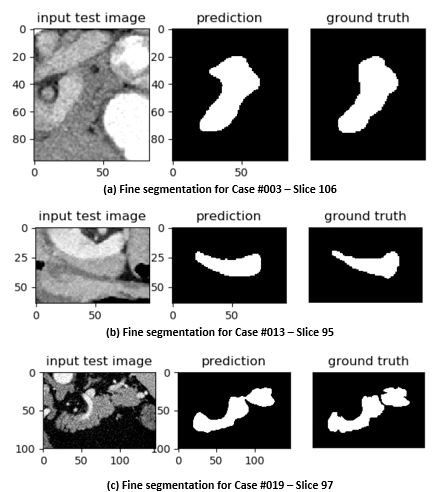 Figure 5. Fine segmentation across multiple cases using R2U-Net.
Figure 5. Fine segmentation across multiple cases using R2U-Net.
Future Work
Combine the coarse-to-fine segmentation model to achieve better accuracy in testing phase. Design a single model to perform the multi-organ segmentation.
References:
1. Roth, H., Lu, L., Farag, A., Shin, H., Liu, J., Turkbey, E., Summers, R.: DeepOrgan: Multi-level Deep Convolutional Networks for Automated Pancreas Segmentation. International Conference on Medical Image Computing and Computer Assisted Intervention (2015)
2. Alom, Md Zahangir, Mahmudul Hasan, Chris Yakopcic, Tarek M. Taha, and Vijayan K. Asari. "Recurrent Residual Convolutional Neural Network based on U-Net (R2U-Net) for Medical Image Segmentation." arXiv preprint arXiv:1802.06955 (2018).
3. Y. Zhou, L. Xie, W. Shen, Y. Wang, E. Fishman and A. Yuille, "A Fixed-Point Model for Pancreas Segmentation in Abdominal CT Scans", Proc. MICCAI, 2017
